The RNA-dependent RNA polymerase (RdRp) drives viral evolution by mediating both genetic drift (mutation) and genetic/antigenic shift (recombination) of RNA viruses. RdRp speed and fidelity contribute to the rate of mutation and formation of the mutant swarm, a well-established determinant of viral fitness and pathogenesis. For more than two decades, we have used the RdRp from poliovirus (PV) as a model system to elucidate fundamental biochemical and biophysical principles governing speed and fidelity of nucleotide addition. A major conclusion of this work is that both speed and fidelity are controlled by the dynamics of a conserved array of amino acid residues in the active site. Importantly, these dynamics can be manipulated genetically or exploited pharmacologically, thus contributing to the creation of attenuated strains and antiviral agents.
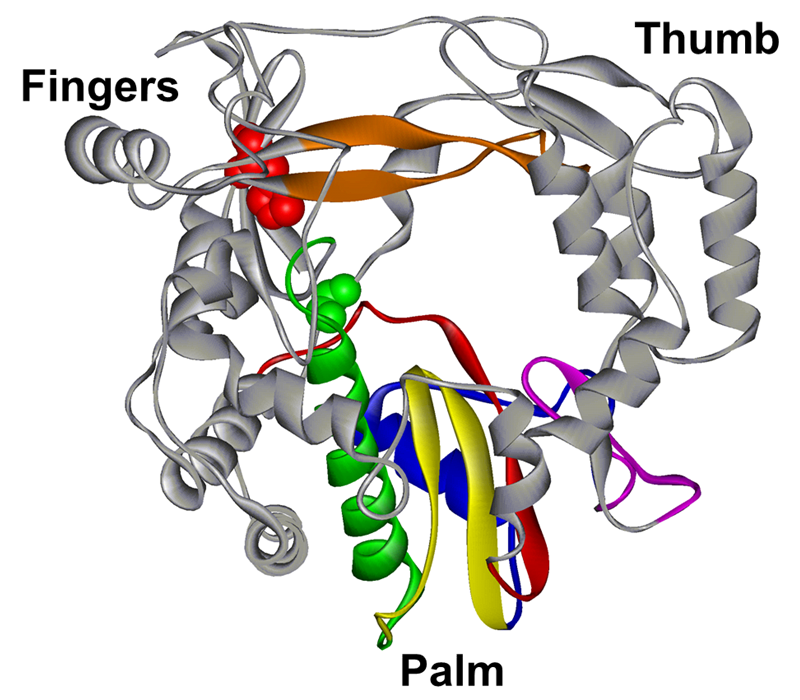
RNA recombination in PV occurs by a template-switching mechanism, a process in which the RdRp initiates elongation on one template (donor) but completes elongation on a different template (acceptor). PV RdRp is sufficient to catalyze template switching in vitro. However, the trigger(s) and mechanism of template switching remain largely unknown. Using a novel cell-based assay for PV RNA recombination, the Evans laboratory observed a direct correlation between RdRp infidelity and the frequency of RNA recombination. RdRp misincorporation frequency as a biochemical property governing template switching was not expected but motivated our foray into the study of RNA recombination. Over the past five years, we have established an experimental paradigm to elucidate the mechanism of RdRp-catalyzed RNA recombination, discover biochemical and biophysical properties and structural determinants of the RdRp governing RNA recombination, and reveal the biological consequences of perturbations to the mechanism and/or efficiency of RNA recombination.
This project uses our experimental paradigm to achieve the following:
- Link structural determinants of the RdRp to elementary steps and mechanisms of RNA recombination;
- Investigate the impact of RNA modifications on elongation and recombination by the RdRp; and
- Reveal the mechanism and biological function of forced-copy-choice RNA recombination.
