Viral infection poses a never-ending threat to human health. Readiness for a viral epidemic of unknown etiology requires broad-spectrum antiviral therapeutics and universal strategies for viral attenuation. Our laboratory has had a longstanding interest in the discovery of knowledge to enable development of approaches to treat and/or to prevent viral infection. We have used poliovirus (PV) as our primary model system to elucidate mechanisms of action of antiviral ribonucleosides with broad-spectrum potential and to reveal RNA-dependent RNA polymerase mechanism-based strategies for viral attenuation.
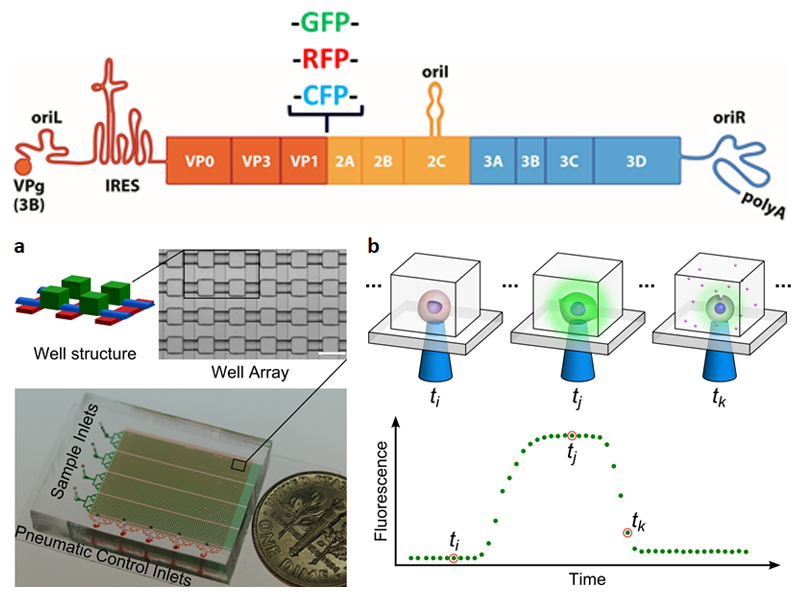
The combination of variation in viral genotype and cellular gene expression predicts variation in the outcomes of infection on the single-cell level in response to antiviral therapies and mechanisms of attenuation. Recently, we developed a microfluidics-based, cell-culturing, imaging and data-analysis platform that enables high-throughput, kinetic analysis of single, isolated cells infected with a PV population expressing enhanced green fluorescent protein. We observed unprecedented, between-cell variation in the onset, speed and yield of replication, as well as variation in lysis, both if and when lysis occurred. Importantly, single-cell analysis revealed the reduced capacity of a PV mutant to establish infection, in part a reflection of a delayed onset of replication. This mutant was attenuated in vivo but without defect by one-step-growth analysis. These studies and those of others demonstrate that single-cell analysis of viral infections can yield knowledge on virus biology and pathogenesis eluded by population methods.
Our current goals are as follows:
- Improve the efficiency of the single-cell experiment;
- Characterize the dynamics of PV translation and genome replication in single cells and determine the extent to which cell type influences these dynamics;
- Determine the extent to which the innate immune system influences the dynamics and outcomes of PV infection within and between cells.
